MET Design
Overview
Multi-environment trials (MET) are experimental designs used to evaluate the performance of treatments across different environments. An environment typically represents a unique combination of location, year, season, or management practice. MET designs are essential for understanding genotype × environment (G×E) interactions, stability, and adaptability of treatments under different conditions.
When to Use
- Treatment evaluation across multiple locations and/or years
- Crop breeding and regional recommendation trials
- Testing stability and adaptability
- When results are intended for large geographic areas
Structure
- Sites: Different locations of the trial
- Site-blocks: Blocks within sites
Setting Up MET Design with speed
Now we can create a data frame representing a MET design. Note that we can specify different dimensions for each site in the designs argument.
all_treatments <- c(rep(1:50, 7), rep(51:57, 8))
met_design <- initialise_design_df(
items = all_treatments,
designs = list(
a = list(nrows = 28, ncols = 5, block_nrows = 7, block_ncols = 5),
b = list(nrows = 20, ncols = 3, block_nrows = 10, block_ncols = 3),
c = list(nrows = 20, ncols = 4, block_nrows = 10, block_ncols = 4),
d = list(nrows = 15, ncols = 4, block_nrows = 5, block_ncols = 4),
e = list(nrows = 22, ncols = 3, block_nrows = 11, block_ncols = 3)
)
)
met_design$site_col <- paste(met_design$site, met_design$col, sep = "_")
met_design$site_block <- paste(met_design$site, met_design$block, sep = "_")
head(met_design) row col treatment row_block col_block block site site_col site_block
1 1 1 1 1 1 1 a a_1 a_1
2 2 1 2 1 1 1 a a_1 a_1
3 3 1 3 1 1 1 a a_1 a_1
4 4 1 4 1 1 1 a a_1 a_1
5 5 1 5 1 1 1 a a_1 a_1
6 6 1 6 1 1 1 a a_1 a_1
Performing the Optimisation
For MET designs, we use lists of named arguments to specify the hierarchical structure. The optimise parameter defines what to optimise and constraints at each level.
optimise <- list(
connectivity = list(spatial_factors = ~site),
balance = list(swap_within = "site", spatial_factors = ~ site_col + site_block)
)
met_result <- speed(
data = met_design,
swap = "treatment",
early_stop_iterations = 5000,
optimise = optimise,
optimise_params = optim_params(random_initialisation = TRUE, adj_weight = 0),
seed = 112
)row and col are used as row and column, respectively.Optimising level: connectivity
Level: connectivity Iteration: 1000 Score: 2.341479 Best: 2.341479 Since Improvement: 26
Level: connectivity Iteration: 2000 Score: 1.091479 Best: 1.091479 Since Improvement: 38
Level: connectivity Iteration: 3000 Score: 0.877193 Best: 0.877193 Since Improvement: 602
Level: connectivity Iteration: 4000 Score: 0.8057644 Best: 0.8057644 Since Improvement: 211
Level: connectivity Iteration: 5000 Score: 0.7700501 Best: 0.7700501 Since Improvement: 169
Level: connectivity Iteration: 6000 Score: 0.7700501 Best: 0.7700501 Since Improvement: 1169
Level: connectivity Iteration: 7000 Score: 0.7700501 Best: 0.7700501 Since Improvement: 2169
Level: connectivity Iteration: 8000 Score: 0.7700501 Best: 0.7700501 Since Improvement: 3169
Level: connectivity Iteration: 9000 Score: 0.7700501 Best: 0.7700501 Since Improvement: 4169
Early stopping at iteration 9831 for level connectivity
Optimising level: balance
Level: balance Iteration: 1000 Score: 8.926692 Best: 8.926692 Since Improvement: 23
Level: balance Iteration: 2000 Score: 7.74812 Best: 7.74812 Since Improvement: 160
Level: balance Iteration: 3000 Score: 7.605263 Best: 7.605263 Since Improvement: 31
Level: balance Iteration: 4000 Score: 7.533835 Best: 7.533835 Since Improvement: 420
Level: balance Iteration: 5000 Score: 7.49812 Best: 7.49812 Since Improvement: 536
Level: balance Iteration: 6000 Score: 7.49812 Best: 7.49812 Since Improvement: 1536
Level: balance Iteration: 7000 Score: 7.49812 Best: 7.49812 Since Improvement: 2536
Level: balance Iteration: 8000 Score: 7.49812 Best: 7.49812 Since Improvement: 3536
Level: balance Iteration: 9000 Score: 7.49812 Best: 7.49812 Since Improvement: 4536
Early stopping at iteration 9464 for level balance
met_resultOptimised Experimental Design
----------------------------
Score: 8.26817
Iterations Run: 19297
Stopped Early: TRUE TRUE
Treatments:
connectivity: 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57
balance: 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57
Seed: 112 Output of the Optimisation
The output shows optimisation results for the design. The score and iterations are combined for the entire design, while the treatments, and stopping criteria are reported separately for each level, allowing you to assess the quality of optimisation at each hierarchy level.
str(met_result)List of 8
$ design_df :'data.frame': 406 obs. of 9 variables:
..$ row : int [1:406] 1 1 1 1 1 1 1 1 1 1 ...
..$ col : int [1:406] 1 1 1 1 1 2 2 2 2 2 ...
..$ treatment : int [1:406] 1 33 3 45 45 49 9 26 9 57 ...
..$ row_block : num [1:406] 1 1 1 1 1 1 1 1 1 1 ...
..$ col_block : num [1:406] 1 1 1 1 1 1 1 1 1 1 ...
..$ block : Factor w/ 4 levels "1","2","3","4": 1 1 1 1 1 1 1 1 1 1 ...
..$ site : chr [1:406] "a" "b" "c" "d" ...
..$ site_col : chr [1:406] "a_1" "b_1" "c_1" "d_1" ...
..$ site_block: chr [1:406] "a_1" "b_1" "c_1" "d_1" ...
$ score : num 8.27
$ scores :List of 2
..$ connectivity: num [1:9832] 5.13 5.2 5.2 5.2 5.2 ...
..$ balance : num [1:9465] 10.3 10.2 10.2 10.3 10.3 ...
$ temperatures :List of 2
..$ connectivity: num [1:9832] 100 99 98 97 96.1 ...
..$ balance : num [1:9465] 100 99 98 97 96.1 ...
$ iterations_run: num 19297
$ stopped_early : Named logi [1:2] TRUE TRUE
..- attr(*, "names")= chr [1:2] "connectivity" "balance"
$ treatments :List of 2
..$ connectivity: chr [1:57] "1" "2" "3" "4" ...
..$ balance : chr [1:57] "1" "2" "3" "4" ...
$ seed : num 112
- attr(*, "class")= chr [1:2] "design" "list"Visualise the Output
ggplot2::ggplot(met_result$design_df, ggplot2::aes(col, row, fill = treatment)) +
ggplot2::geom_tile(color = "black") +
ggplot2::scale_fill_viridis_c() +
ggplot2::facet_wrap(~site, scales = "free") +
ggplot2::scale_x_continuous(expand = c(0, 0), breaks = 1:max(met_design$col)) +
ggplot2::scale_y_continuous(expand = c(0, 0), breaks = 1:max(met_design$row), trans = scales::reverse_trans()) +
ggplot2::theme_bw() +
ggplot2::theme(
legend.position = "none",
panel.grid.major = ggplot2::element_blank(),
panel.grid.minor = ggplot2::element_blank()
)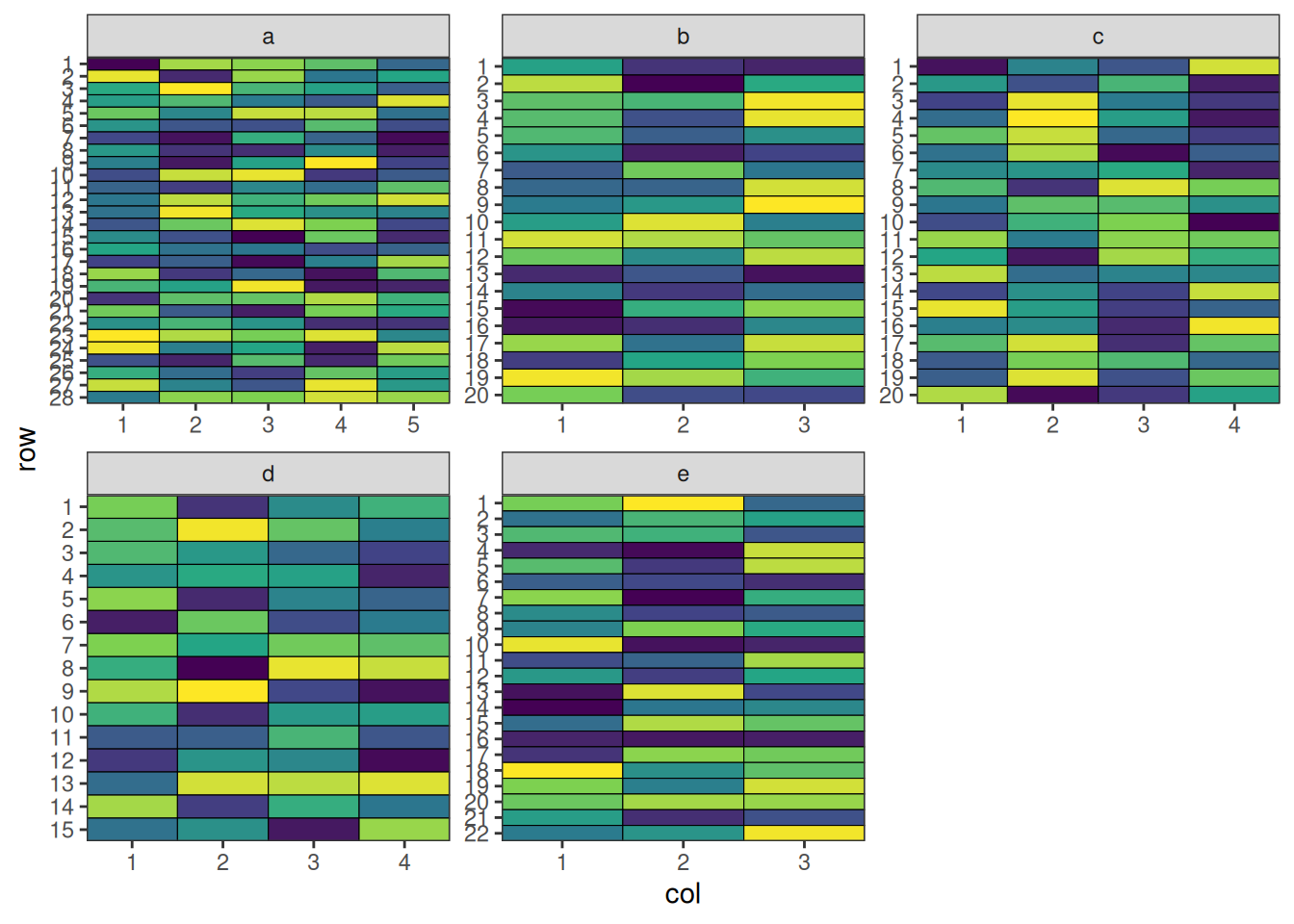
This design has now been optimised at both the connectivity between sites and the balance within each site.
Spatial Design Considerations
Field Shape and Orientation
Neighbour Effects
Using speed Effectively
- Set appropriate parameters: Balance optimisation time with improvement
- Visualise designs: Always plot layouts before implementation
- Compare alternatives: Test multiple blocking strategies
- Validate results: Check constraint satisfaction and efficiency metrics
Conclusion
Further Reading
Related Vignettes
This vignette demonstrates the versatility of the speed package for agricultural experimental design. For more advanced applications and custom designs, consult the package documentation and additional vignettes.